
We study the molecular processes that underlie the epigenetic regulation of gene expression and transposable element activity.
Gene silencing refers to the repression of genes through epigenetic mechanisms, without altering the DNA sequence. In both plants and animals, gene silencing is tightly associated with modifications to chromatin, such as cytosine DNA methylation and particular post-translational modification of histone proteins. Gene silencing does not only affect exogenous DNA entering the genome, such as viral sequences, but also endogenous genomic sequences such as certain protein-coding genes and most transposons. Efficient and accurate gene silencing is therefore essential for proper gene expression and genome stability. Our research combines genetic and genomic approaches to understand the mechanisms of gene silencing, primarily using the flowering plant Arabidopsis as a model.
Funders

Research

 In eukaryotic organisms, the genome is packaged in the form of chromatin, which is composed of DNA molecules and various proteins, including histones. The structure of chromatin, including its chemical composition and degree of compaction, plays a critical role in the regulation of gene expression. This structure is influenced by the types of proteins present, such as different histone variants, as well as by chemical modifications, known as “epigenetic marks,” on both the DNA and histones.
In eukaryotic organisms, the genome is packaged in the form of chromatin, which is composed of DNA molecules and various proteins, including histones. The structure of chromatin, including its chemical composition and degree of compaction, plays a critical role in the regulation of gene expression. This structure is influenced by the types of proteins present, such as different histone variants, as well as by chemical modifications, known as “epigenetic marks,” on both the DNA and histones.
Our research focuses on understanding the molecular mechanisms of epigenetic silencing. We use genetic and genomic approaches to analyze the pathways controlling the genomic distribution of different epigenetic marks and their interactions. Additionally, we study the molecular pathways involved in silencing transposable elements and certain genes, with a particular interest in DNA methylation-independent silencing mechanisms. Our goal is to uncover the fundamental mechanisms that control gene expression and genome stability through chromatin structure and epigenetic regulation.
Alumni
PhD Students
Amy Hesketh (2017-2021)
Pierre Bourguet (2014-2018)
Maxime Auzon-Cape (2013-2017)
Mélanie Rigal (2010-2014)
Postdoctoral fellows
Sébastien Lageix (2014-2015)
Romain Pogorelcnik (2013-2015)
Angeles Gomez Zambrano (2013-2015)
Leticia Lopez Gonzalez (2012-2014)
Stève de Bossoreille de Ribou (2012-2014)
Anthony Devert (2012-2013)
Yoko Ikeda (2011-2013)
Zoltan Kevei (2010-2013)
Members
Publications
Plant Mobile Domain protein-DNA motif modules counteract Polycomb silencing to stabilize gene expression
Published on 28 Sep 2024 in BioRxiv
Pelissier T , Jarry L, Olivier M , Dajoux G , Pouch-Pelissier MN , Courtois C , Descombin J, Picault N, Moissiard G, Mathieu O
Genetic-epigenetic interplay in the determination of plant 3D genome organization.
Published on 23 Sep 2024 in Nucleic acids research , vol. 52 - pp 10220-10234
He X, Dias Lopes C, Pereyra-Bistrain LI, Huang Y, An J, Chaouche RB, Zalzalé H, Wang Q, Ma X, Antunez-Sanchez J, Bergounioux C, Piquerez S, Fragkostefanakis S, Zhang Y, Zheng S, Crespi M, Mahfouz MM, Mathieu O , Ariel F, Gutierrez-Marcos J, Li X, Bouché N, Raynaud C, Latrasse D, Benhamed M
An atlas of the tomato epigenome reveals that KRYPTONITE shapes TAD-like boundaries through the control of H3K9ac distribution.
Published on 09 Jul 2024 in Proceedings of the National Academy of Sciences of the United States of America , vol. 121 - pp e2400737121
An J, Brik Chaouche R, Pereyra-Bistraín LI, Zalzalé H, Wang Q, Huang Y, He X, Dias Lopes C, Antunez-Sanchez J, Bergounioux C, Boulogne C, Dupas C, Gillet C, Pérez-Pérez JM, Mathieu O , Bouché N, Fragkostefanakis S, Zhang Y, Zheng S, Crespi M, Mahfouz MM, Ariel F, Gutierrez-Marcos J, Raynaud C, Latrasse D, Benhamed M
RTEL1 is required for silencing and epigenome stability.
Published on 08 Sep 2023 in Nucleic acids research , vol. 51 - pp 8463-8479
Olivier M , Hesketh A , Pouch-Pélissier MN , Pélissier T , Huang Y, Latrasse D, Benhamed M, Mathieu O
Reaching the inaccessible DNA.
Published on 30 Jun 2022 in Nature reviews. Molecular cell biology , vol. 23 - pp 388
Crosstalk between H2A variant-specific modifications impacts vital cell functions.
Published on 30 Jun 2021 in PLoS genetics , vol. 17 - pp e1009601
Schmücker A, Lei B, Lorković ZJ, Capella M, Braun S, Bourguet P , Mathieu O , Mechtler K, Berger F
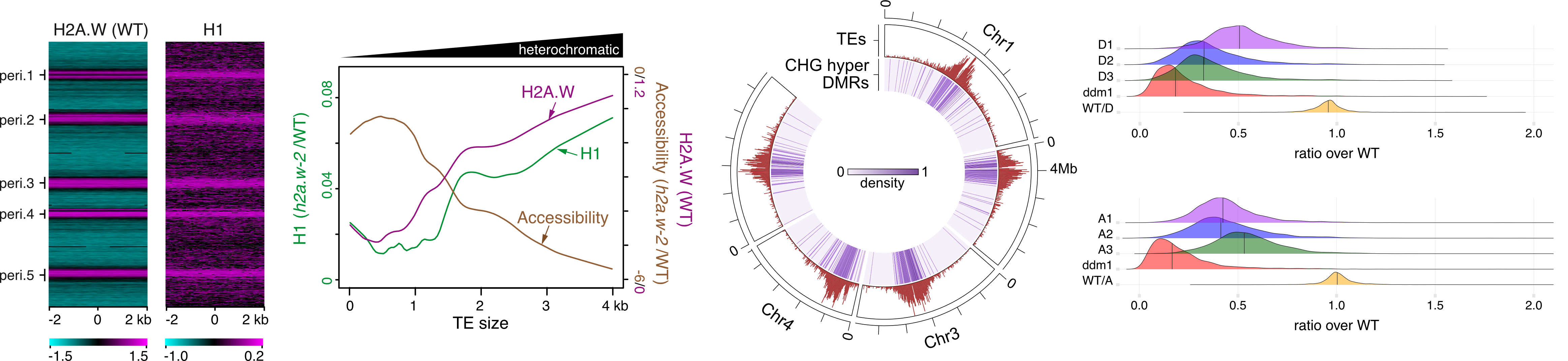
The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation.
Published on 11 May 2021 in Nature communications , vol. 12 - pp 2683
Bourguet P , Picard CL, Yelagandula R, Pélissier T , Lorković ZJ, Feng S, Pouch-Pélissier MN , Schmücker A, Jacobsen SE, Berger F, Mathieu O
DNA polymerase epsilon is required for heterochromatin maintenance in Arabidopsis.
Published on 25 Nov 2020 in Genome biology , vol. 21 - pp 283
Bourguet P , López-González L, Gómez-Zambrano Á, Pélissier T , Hesketh A , Potok ME, Pouch-Pélissier MN , Perez M, Da Ines O , Latrasse D, White CI , Jacobsen SE, Benhamed M, Mathieu O
PP7L is essential for MAIL1-mediated transposable element silencing and primary root growth.
Published on 30 May 2020 in The Plant journal : for cell and molecular biology , vol. 102 - pp 703-717
de Luxán-Hernández C, Lohmann J, Hellmeyer W, Seanpong S, Wöltje K, Magyar Z, Pettkó-Szandtner A, Pélissier T , De Jaeger G, Hoth S, Mathieu O , Weingartner M
A role for MED14 and UVH6 in heterochromatin transcription upon destabilization of silencing.
Published on 30 Dec 2018 in Life science alliance , vol. 1 - pp e201800197
Bourguet P , de Bossoreille S, López-González L, Pouch-Pélissier MN , Gómez-Zambrano Á, Devert A , Pélissier T , Pogorelcnik R , Vaillant I , Mathieu O
Loss of CG Methylation in Marchantia polymorpha Causes Disorganization of Cell Division and Reveals Unique DNA Methylation Regulatory Mechanisms of Non-CG Methylation.
Published on 01 Dec 2018 in Plant & cell physiology , vol. 59 - pp 2421-2431
Ikeda Y, Nishihama R, Yamaoka S, Arteaga-Vazquez MA, Aguilar-Cruz A, Grimanelli D, Pogorelcnik R, Martienssen RA, Yamato KT, Kohchi T, Hirayama T, Mathieu O
Arabidopsis proteins with a transposon-related domain act in gene silencing.
Published on 03 May 2017 in Nature communications , vol. 8 - pp 15122
Ikeda Y, Pélissier T , Bourguet P , Becker C, Pouch-Pélissier MN , Pogorelcnik R , Weingartner M, Weigel D, Deragon JM, Mathieu O
Evolutionary history of double-stranded RNA binding proteins in plants: identification of new cofactors involved in easiRNA biogenesis.
Published on 30 May 2016 in Plant molecular biology , vol. 91 - pp 131-47
Clavel M, Pélissier T , Montavon T, Tschopp MA, Pouch-Pélissier MN , Descombin J, Jean V, Dunoyer P, Bousquet-Antonelli C, Deragon JM
Epigenome confrontation triggers immediate reprogramming of DNA methylation and transposon silencing in Arabidopsis thaliana F1 epihybrids.
Published on 05 Apr 2016 in Proceedings of the National Academy of Sciences of the United States of America , vol. 113 - pp E2083-92
Rigal M , Becker C, Pélissier T , Pogorelcnik R , Devos J, Ikeda Y, Weigel D, Mathieu O
Parallel action of AtDRB2 and RdDM in the control of transposable element expression.
Published on 03 Mar 2015 in BMC plant biology , vol. 15 - pp 70
Clavel M, Pélissier T , Descombin J, Jean V, Picart C, Charbonel C, Saez-Vásquez J, Bousquet-Antonelli C, Deragon JM
Interplay between chromatin and RNA processing.
Published on 30 Apr 2014 in Current opinion in plant biology , vol. 18 - pp 60-5
Mathieu O , Bouché N
DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns.
Published on 29 Jun 2012 in The EMBO journal , vol. 31 - pp 2981-93
Rigal M , Kevei Z , Pélissier T , Mathieu O
Double-stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV-dependent siRNA levels in Arabidopsis.
Published on 30 Aug 2011 in RNA (New York, N.Y.) , vol. 17 - pp 1502-10
Pélissier T , Clavel M, Chaparro C, Pouch-Pélissier MN , Vaucheret H, Deragon JM
A “mille-feuille” of silencing: epigenetic control of transposable elements.
Published on 12 Aug 2011 in Biochimica et biophysica acta , vol. 1809 - pp 452-8
Rigal M, Mathieu O
Stress-induced activation of heterochromatic transcription.
Published on 28 Oct 2010 in PLoS genetics , vol. 6 - pp e1001175
Tittel-Elmer M, Bucher E, Broger L, Mathieu O , Paszkowski J, Vaillant I
Bursts of retrotransposition reproduced in Arabidopsis.
Published on 17 Sep 2009 in Nature , vol. 461 - pp 423-6
Tsukahara S, Kobayashi A, Kawabe A, Mathieu O , Miura A, Kakutani T
Selective epigenetic control of retrotransposition in Arabidopsis.
Published on 17 Sep 2009 in Nature , vol. 461 - pp 427-30
Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O
Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding.
Published on 24 Dec 2008 in BMC plant biology , vol. 8 - pp 134
Lageix S , Lanet E, Pouch-Pélissier MN , Espagnol MC , Robaglia C, Deragon JM, Pélissier T
Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants.
Published on 22 Aug 2008 in PLoS genetics , vol. 4 - pp e1000165
Caikovski M, Yokthongwattana C, Habu Y, Nishimura T, Mathieu O , Paszkowski J
SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex.
Published on 13 Jun 2008 in PLoS genetics , vol. 4 - pp e1000096
Pouch-Pélissier MN , Pélissier T , Elmayan T, Vaucheret H, Boko D, Jantsch MF, Deragon JM















