We study the epigenetic basis of cellular identity in mammals.
The differentiation of a cell into a specialized cell type is a complex process that requires the coordinated action of multiple regulatory layers to establish a new expression program. However, our understanding of the machinery orchestrating this process, as well as the causes and consequences of its disruption, remains incomplete.
Our research aims to identify the mechanisms that coordinate different levels of epigenetic regulation during the acquisition of neural identity and to understand their deregulation in pathological contexts.
To this end, we use genomic imprinting and malignant glioma as model systems.
Funders

Research
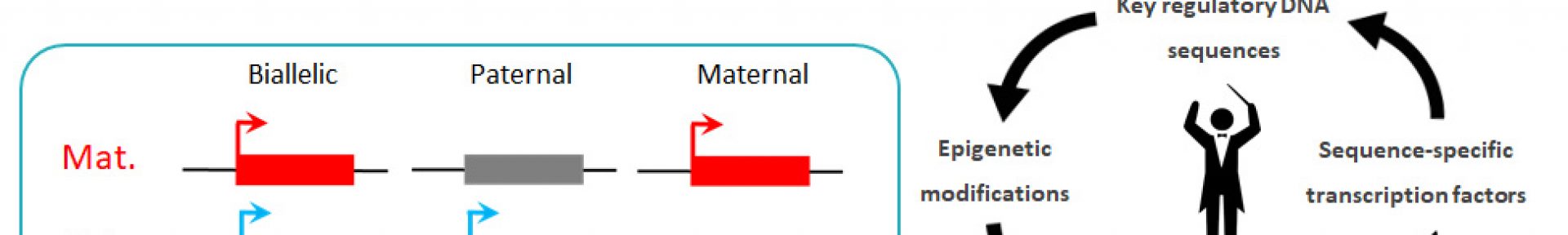
Genomic Imprinting : From regulation to Function
Regulation of Imprinting Loci in Neural Lineages (Contacts: F. Court, P. Arnaud)
Genomic imprinting restricts the expression of nearly 150 mammalian genes to a single parental allele. These genes play key roles in growth control and brain function. At the molecular level, they are organized into about twenty genomic domains, each regulated by an imprinting control region (ICR) and the coordinated action of multiple regulatory layers. However, the mechanisms that orchestrate this multi-scale regulation and enable the ICR to remotely control the monoallelic expression of genes during cellular identity acquisition remain poorly understood. Our goal is to identify these mechanisms and the key players involved in neural identity acquisition.
To achieve this, we need to simultaneously assess the dynamics of multiple regulatory levels during cellular identity acquisition. To this end, we have developed an integrated approach that combines multi-scale experimental studies with functional analyses using 2D and 3D (organoids) in vitro corticogenesis systems that recapitulate imprinting patterns observed in the early (Bouschet et al., Cereb Cortex, 2017) and postnatal brain, respectively. This animal-reduced system relies on the controlled differentiation of reciprocal hybrid murine embryonic stem (ES) cells, enabling the dynamic and allelic study of molecular signatures.
Our strategy integrates comprehensive molecular (DNA methylation, gene expression, histone modifications, and regulatory factors), 3D chromatin conformation, and bioinformatics analyses. This has allowed us to generate a high-resolution map providing an integrative, allelic, and dynamic view of both the linear molecular signatures and 3D chromatin organization associated with ICR regions during murine neural lineage formation. This framework uniquely enables us to dissect the regulatory mechanisms of multiple imprinting loci, which we are functionally testing using CRISPR-based approaches.
Function of Imprinting Loci: The H13/Mcts2 Locus (Contacts: B. Montibus,P. Arnaud)
Disruptions in the transcriptional regulation of genomic imprinting, including alterations in allele-specific isoform production, can lead to imprinting disorders (IDs)—a group of rare congenital conditions affecting growth and neurodevelopment. Identifying the genetic causes and pathophysiological mechanisms underlying IDs remains a major challenge.
A striking example is Mulchandani–Bhoj–Conlin syndrome (MBCS), a disorder characterized by fetal and postnatal growth restriction, short stature, failure to thrive, and developmental delay. Evidence suggests that dysregulated expression of the imprinted HM13/Mcts2 locus contributes to MBCS. To test this hypothesis, our project aims to elucidate the regulatory mechanisms and functional roles of the H13/Mcts2 locus to assess its impact on MBCS pathogenesis.
To achieve this, we employ a combination of phenotypic analyses and molecular studies at both the transcriptional and translational levels in mouse models and cell lines. Our approach integrates descriptive and functional studies to uncover the roles of allele-specific and isoform expression during development and the consequences of their deregulation in disease.
Glioma: study of chimeric transcripts from LINE L1 elements
(Contacts: C. Vaurs-Barriére; P Arnaud)
This project was initiated to identify the mechanisms that coordinate different levels of epigenetic regulation by investigating the causes and consequences of their alteration in malignant glioma, one of the most common types of brain tumors. Collaborations with clinical teams allow us to carry out this study on samples from the Auvergne Glioma Tumorotheque and glioblastoma initiating cell lines (GIC). An exhaustive molecular study allowed us to reveal that the majority of transcriptional defects in gliomas are not related to DNA methylation defects but rather due to an alteration in the control of H3K27me3 (here). The search for the causes of this alteration leads us to study in more detail the (de)regulation of HOX genes, for which we have established that the alteration is a signature of the most aggressive gliomas.
To identify new candidates we have undertaken, in a parallel approach, to explore the link between deregulation of transposable elements and alteration of H3K27me3 in cancer cells. Our original hypothesis is that antisense transcription from L1 repeats can induce aberrant expression of adjacent genes, in the form of so-called LINE-1 chimeric transcripts (LCTs), which in turn influence the dynamics of the H3K27me3 mark. We have previously developed the CLIFinder bioinformatics tool to identify these chimeric transcripts from RNA seq data (here). With this tool, we have, for the first time, established the panorama of LCTs in aggressive gliomas and undertaken to characterise the altered genes in CIGs.
Members
Publications
PIK3R1 and G0S2 are human placenta-specific imprinted genes associated with germline-inherited maternal DNA methylation.
Published on 01 Dec 2025 in Epigenetics , vol. 20 - pp 2523191
Daskeviciute D, Sainty B, Chappell-Maor L, Bone C, Russell S, Iglesias-Platas I, Arnaud P , Monteagudo-Sánchez A, Greenberg MVC, Chen K, Manerao-Azua A, Perez de Nanclares G, Lartey J, Monk D
Non-canonical imprinting, manifesting as post-fertilization placenta-specific parent-of-origin dependent methylation, is not conserved in humans.
Published on 17 Jan 2025 in Human molecular genetics
Daskeviciute D, Chappell-Maor L, Sainty B, Arnaud P , Iglesias-Platas I, Simon C, Okae H, Arima T, Vassena R, Lartey J, Monk D
Biallelic non-productive enhancer-promoter interactions precede imprinted expression of Kcnk9 during mouse neural commitment.
Published on 30 Jan 2024 in HGG advances , vol. 5 - pp 100271
Rengifo Rojas C , Cercy J , Perillous S , Gonthier-Guéret C, Montibus B , Maupetit-Méhouas S, Espinadel A , Dupré M , Hong CC, Hata K, Nakabayashi K, Plagge A, Bouschet T, Arnaud P , Vaillant I , Court F
Variable allelic expression of imprinted genes at the Peg13, Trappc9, Ago2 cluster in single neural cells.
Published on 12 Oct 2022 in Frontiers in cell and developmental biology , vol. 10 - pp 1022422
Claxton M, Pulix M, Seah MKY, Bernardo R, Zhou P, Aljuraysi S, Liloglou T, Arnaud P , Kelsey G, Messerschmidt DM, Plagge A
L1 chimeric transcripts are expressed in healthy brain and their deregulation in glioma follows that of their host locus.
Published on 17 Aug 2022 in Human molecular genetics , vol. 31 - pp 2606-2622
Pinson ME , Court F , Masson A , Renaud Y , Fantini A , Bacoeur-Ouzillou O, Barriere M, Pereira B, Guichet PO, Chautard E, Karayan-Tapon L, Verrelle P, Arnaud P , Vaurs-Barrière C
The Long Non-Coding RNA HOXA-AS2 Promotes Proliferation of Glioma Stem Cells and Modulates Their Inflammation Pathway Mainly through Post-Transcriptional Regulation.
Published on 25 Apr 2022 in International journal of molecular sciences , vol. 23
Le Boiteux E , Guichet PO, Masliantsev K, Montibus B , Vaurs-Barriere C, Gonthier-Gueret C, Chautard E, Verrelle P, Karayan-Tapon L, Fogli A , Court F , Arnaud P
Widespread overexpression from the four DNA hypermethylated HOX clusters in aggressive (IDHwt) glioma is associated with H3K27me3 depletion and alternative promoter usage.
Published on 30 Aug 2021 in Molecular oncology , vol. 15 - pp 1995-2010
Le Boiteux E , Court F , Guichet PO, Vaurs-Barrière C , Vaillant I , Chautard E, Verrelle P, Costa BM, Karayan-Tapon L, Fogli A , Arnaud P
TET3 controls the expression of the H3K27me3 demethylase Kdm6b during neural commitment.
Published on 30 Jan 2021 in Cellular and molecular life sciences : CMLS , vol. 78 - pp 757-768
Montibus B , Cercy J , Bouschet T, Charras A, Maupetit-Méhouas S , Nury D , Gonthier-Guéret C, Chauveau S , Allegre N , Chariau C, Hong CC, Vaillant I , Marques CJ, Court F , Arnaud P
HOX gene cluster (de)regulation in brain: from neurodevelopment to malignant glial tumours.
Published on 30 Oct 2020 in Cellular and molecular life sciences : CMLS , vol. 77 - pp 3797-3821
Gonçalves CS, Le Boiteux E , Arnaud P , Costa BM
Transcriptional alterations in glioma result primarily from DNA methylation-independent mechanisms.
Published on 30 Oct 2019 in Genome research , vol. 29 - pp 1605-1621
Court F , Le Boiteux E , Fogli A , Müller-Barthélémy M, Vaurs-Barrière C , Chautard E, Pereira B, Biau J, Kemeny JL, Khalil T, Karayan-Tapon L, Verrelle P, Arnaud P
Childhood Ataxia with Central Nervous System Hypomyelination / Vanishing White Matter.
Published on 04 Apr 2019
Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, van der Knaap MS, Fogli A , Boespflug-Tanguy O, Abbink TEM, Schiffmann R
High-salt-recovered sequences are associated with the active chromosomal compartment and with large ribonucleoprotein complexes including nuclear bodies.
Published on 30 Nov 2018 in Genome research , vol. 28 - pp 1733-1746
Baudement MO, Cournac A, Court F , Seveno M, Parrinello H, Reynes C, Sabatier R, Bouschet T, Yi Z, Sallis S, Tancelin M, Rebouissou C, Cathala G, Lesne A, Mozziconacci J, Journot L, Forné T
Oxidative stress and mitochondrial dynamics malfunction are linked in Pelizaeus-Merzbacher disease.
Published on 30 Sep 2018 in Brain pathology (Zurich, Switzerland) , vol. 28 - pp 611-630
Ruiz M, Bégou M, Launay N, Ranea-Robles P, Bianchi P, López-Erauskin J, Morató L, Guilera C, Petit B, Vaurs-Barriere C , Guéret-Gonthier C, Bonnet-Dupeyron MN, Fourcade S, Auwerx J, Boespflug-Tanguy O, Pujol A
DNA methylation profiling reveals a pathological signature that contributes to transcriptional defects of CD34(+) CD15(-) cells in early chronic-phase chronic myeloid leukemia.
Published on 30 Jun 2018 in Molecular oncology , vol. 12 - pp 814-829
Maupetit-Mehouas S , Court F , Bourgne C, Guerci-Bresler A, Cony-Makhoul P, Johnson H, Etienne G, Rousselot P, Guyotat D, Janel A, Hermet E, Saugues S, Berger J, Arnaud P , Berger MG
The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma.
Published on 20 Mar 2018 in Oncotarget , vol. 9 - pp 15740-15756
Xavier-Magalhães A, Gonçalves CS, Fogli A , Lourenço T, Pojo M, Pereira B, Rocha M, Lopes MC, Crespo I, Rebelo O, Tão H, Lima J, Moreira R, Pinto AA, Jones C, Reis RM, Costello JF, Arnaud P , Sousa N, Costa BM
CLIFinder: identification of LINE-1 chimeric transcripts in RNA-seq data.
Published on 15 Feb 2018 in Bioinformatics (Oxford, England) , vol. 34 - pp 688-690
Pinson ME , Pogorelcnik R , Court F , Arnaud P , Vaurs-Barrière C
Recommendations for a nomenclature system for reporting methylation aberrations in imprinted domains.
Published on 01 Jan 2018 in Epigenetics , vol. 13 - pp 117-121
Monk D, Morales J, den Dunnen JT, Russo S, Court F , Prawitt D, Eggermann T, Beygo J, Buiting K, Tümer Z, Nomenclature group of the European Network for Human Congenital Imprinting Disorders
Detection of the alternative lengthening of telomeres pathway in malignant gliomas for improved molecular diagnosis.
Published on 30 Nov 2017 in Journal of neuro-oncology , vol. 135 - pp 381-390
Fogli A , Demattei MV, Corset L, Vaurs-Barrière C , Chautard E, Biau J, Kémény JL, Godfraind C, Pereira B, Khalil T, Grandin N , Arnaud P , Charbonneau M , Verrelle P
Maternal mutations of FOXF1 cause alveolar capillary dysplasia despite not being imprinted.
Published on 30 Jun 2017 in Human mutation , vol. 38 - pp 615-620
Alsina Casanova M, Monteagudo-Sánchez A, Rodiguez Guerineau L, Court F , Gazquez Serrano I, Martorell L, Rovira Zurriaga C, Moore GE, Ishida M, Castañon M, Moliner Calderon E, Monk D, Moreno Hernando J
In Vitro Corticogenesis from Embryonic Stem Cells Recapitulates the In Vivo Epigenetic Control of Imprinted Gene Expression.
Published on 01 Mar 2017 in Cerebral cortex (New York, N.Y. : 1991) , vol. 27 - pp 2418-2433
Bouschet T, Dubois E, Reynès C, Kota SK, Rialle S, Maupetit-Méhouas S , Pezet M, Le Digarcher A, Nidelet S, Demolombe V, Cavelier P, Meusnier C, Maurizy C, Sabatier R, Feil R, Arnaud P , Journot L, Varrault A
DNA methylation profiling identifies PTRF/Cavin-1 as a novel tumor suppressor in Ewing sarcoma when co-expressed with caveolin-1.
Published on 01 Feb 2017 in Cancer letters , vol. 386 - pp 196-207
Huertas-Martínez J, Court F , Rello-Varona S, Herrero-Martín D, Almacellas-Rabaiget O, Sáinz-Jaspeado M, Garcia-Monclús S, Lagares-Tena L, Buj R, Hontecillas-Prieto L, Sastre A, Azorin D, Sanjuan X, López-Alemany R, Moran S, Roma J, Gallego S, Mora J, García Del Muro X, Giangrande PH, Peinado MA, Alonso J, de Alava E, Monk D, Esteller M, Tirado OM
An annotated list of bivalent chromatin regions in human ES cells: a new tool for cancer epigenetic research.
Published on 17 Jan 2017 in Oncotarget , vol. 8 - pp 4110-4124
Quantitative Analysis of Intra-chromosomal Contacts: The 3C-qPCR Method.
Published on 01 Jan 2017 in Methods in molecular biology (Clifton, N.J.) , vol. 1589 - pp 75-88
Ea V, Court F , Forné T
Human Oocyte-Derived Methylation Differences Persist in the Placenta Revealing Widespread Transient Imprinting.
Published on 30 Nov 2016 in PLoS genetics , vol. 12 - pp e1006427
Sanchez-Delgado M, Court F , Vidal E, Medrano J, Monteagudo-Sánchez A, Martin-Trujillo A, Tayama C, Iglesias-Platas I, Kondova I, Bontrop R, Poo-Llanillo ME, Marques-Bonet T, Nakabayashi K, Simón C, Monk D
The tumoral A genotype of the MGMT rs34180180 single-nucleotide polymorphism in aggressive gliomas is associated with shorter patients’ survival.
Published on 01 Mar 2016 in Carcinogenesis , vol. 37 - pp 169-176
Fogli A , Chautard E, Vaurs-Barrière C , Pereira B, Müller-Barthélémy M, Court F , Biau J, Pinto AA, Kémény JL, Khalil T, Karayan-Tapon L, Verrelle P, Costa BM, Arnaud P
Imprinting control regions (ICRs) are marked by mono-allelic bivalent chromatin when transcriptionally inactive.
Published on 29 Jan 2016 in Nucleic acids research , vol. 44 - pp 621-35
Maupetit-Méhouas S , Montibus B , Nury D , Tayama C, Wassef M, Kota SK, Fogli A , Cerqueira Campos F , Hata K, Feil R, Margueron R, Nakabayashi K, Court F , Arnaud P
Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape.
Published on 25 Sep 2015 in Genome biology , vol. 16 - pp 209
Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S , Arnaud P , Tomizawa S, Andrews S, Kelsey G
ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain.
Published on 13 Oct 2014 in Developmental cell , vol. 31 - pp 19-33
Kota SK, Llères D, Bouschet T, Hirasawa R, Marchand A, Begon-Pescia C, Sanli I, Arnaud P , Journot L, Girardot M, Feil R
Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer.
Published on 30 May 2014 in Epigenetics , vol. 9 - pp 783-90
Romanelli V, Nakabayashi K, Vizoso M, Moran S, Iglesias-Platas I, Sugahara N, Simón C, Hata K, Esteller M, Court F , Monk D
Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment.
Published on 30 Apr 2014 in Genome research , vol. 24 - pp 554-69
Court F , Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simón C, Moore H, Harness JV, Keirstead H, Sanchez-Mut JV, Kaneki E, Lapunzina P, Soejima H, Wake N, Esteller M, Ogata T, Hata K, Nakabayashi K, Monk D
The PEG13-DMR and brain-specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus.
Published on 25 Mar 2014 in Epigenetics & chromatin , vol. 7 - pp 5
Court F , Camprubi C, Garcia CV, Guillaumet-Adkins A, Sparago A, Seruggia D, Sandoval J, Esteller M, Martin-Trujillo A, Riccio A, Montoliu L, Monk D
Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing.
Published on 20 Feb 2014 in Molecular cell , vol. 53 - pp 672-81
Liang J, Lacroix L, Gamot A, Cuddapah S, Queille S, Lhoumaud P, Lepetit P, Martin PG, Vogelmann J, Court F , Hennion M, Micas G, Urbach S, Bouchez O, Nöllmann M, Zhao K, Emberly E, Cuvier O
Hypermethylation of the alternative AWT1 promoter in hematological malignancies is a highly specific marker for acute myeloid leukemias despite high expression levels.
Published on 09 Jan 2014 in Journal of hematology & oncology , vol. 7 - pp 4
Guillaumet-Adkins A, Richter J, Odero MD, Sandoval J, Agirre X, Catala A, Esteller M, Prósper F, Calasanz MJ, Buño I, Kwon M, Court F , Siebert R, Monk D
Comparison of three methods of diagnosis of plasma unmeasured anions in critically ill patients.
Published on 30 Oct 2013 in Minerva anestesiologica , vol. 79 - pp 1164-72
Lautrette A, Fejjal M, Aithssain A, Phan TN, Caillot N, Fogli A , Souweine B
Chromatin loop organization of the junb locus in mouse dendritic cells.
Published on 30 Oct 2013 in Nucleic acids research , vol. 41 - pp 8908-25
Salem T, Gomard T, Court F , Moquet-Torcy G, Brockly F, Forné T, Piechaczyk M
Stability of genomic imprinting and gestational-age dynamic methylation in complicated pregnancies conceived following assisted reproductive technologies.
Published on 30 Sep 2013 in Biology of reproduction , vol. 89 - pp 50
Camprubí C, Iglesias-Platas I, Martin-Trujillo A, Salvador-Alarcon C, Rodriguez MA, Barredo DR, Court F , Monk D
Liver x receptors protect from development of prostatic intra-epithelial neoplasia in mice.
Published on 30 May 2013 in PLoS genetics , vol. 9 - pp e1003483
Pommier AJ, Dufour J , Alves G, Viennois E, De Boussac H , Trousson A , Volle DH , Caira F , Val P , Arnaud P , Lobaccaro JM , Baron S
Genome-wide allelic methylation analysis reveals disease-specific susceptibility to multiple methylation defects in imprinting syndromes.
Published on 30 Apr 2013 in Human mutation , vol. 34 - pp 595-602
Court F , Martin-Trujillo A, Romanelli V, Garin I, Iglesias-Platas I, Salafsky I, Guitart M, Perez de Nanclares G, Lapunzina P, Monk D
Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse.
Published on 30 Mar 2013 in Development (Cambridge, England) , vol. 140 - pp 1231-9
Borensztein M, Monnier P, Court F , Louault Y, Ripoche MA, Tiret L, Yao Z, Tapscott SJ, Forné T, Montarras D, Dandolo L
Imprinting at the PLAGL1 domain is contained within a 70-kb CTCF/cohesin-mediated non-allelic chromatin loop.
Published on 01 Feb 2013 in Nucleic acids research , vol. 41 - pp 2171-9
Iglesias-Platas I, Court F , Camprubi C, Sparago A, Guillaumet-Adkins A, Martin-Trujillo A, Riccio A, Moore GE, Monk D
A yeast purification system for human translation initiation factors eIF2 and eIF2Bε and their use in the diagnosis of CACH/VWM disease.
Published on 01 Jan 2013 in PloS one , vol. 8 - pp e53958
de Almeida RA, Fogli A , Gaillard M, Scheper GC, Boesflug-Tanguy O, Pavitt GD
Characterization of novel paternal ncRNAs at the Plagl1 locus, including Hymai, predicted to interact with regulators of active chromatin.
Published on 19 Jun 2012 in PloS one , vol. 7 - pp e38907
Iglesias-Platas I, Martin-Trujillo A, Cirillo D, Court F , Guillaumet-Adkins A, Camprubi C, Bourc'his D, Hata K, Feil R, Tartaglia G, Arnaud P , Monk D
Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.
Published on 01 Feb 2012 in The EMBO journal , vol. 31 - pp 606-15
Henckel A, Chebli K, Kota SK, Arnaud P , Feil R
Relevance of GJC2 promoter mutation in Pelizaeus-Merzbacher-like disease.
Published on 30 Jan 2012 in Annals of neurology , vol. 71 - pp 146-8
Combes P, Kammoun N, Monnier A, Gonthier-Guéret C, Giraud G , Bertini E, Chahnez T, Fakhfakh F, Boespflug-Tanguy O, Vaurs-Barrière C
Sjögren-Larsson syndrome: novel mutations in the ALDH3A2 gene in a French cohort.
Published on 15 Jan 2012 in Journal of the neurological sciences , vol. 312 - pp 123-6
Sarret C, Rigal M , Vaurs-Barrière C , Dorboz I, Eymard-Pierre E, Combes P, Giraud G , Wanders RJ, Afenjar A, Francannet C, Boespflug-Tanguy O
H19 antisense RNA can up-regulate Igf2 transcription by activation of a novel promoter in mouse myoblasts.
Published on 01 Jan 2012 in PloS one , vol. 7 - pp e37923
Tran VG, Court F , Duputié A, Antoine E, Aptel N, Milligan L, Carbonell F, Lelay-Taha MN, Piette J, Weber M, Montarras D, Pinset C, Dandolo L, Forné T, Cathala G
CSF N-glycan profiles to investigate biomarkers in brain developmental disorders: application to leukodystrophies related to eIF2B mutations.
Published on 01 Jan 2012 in PloS one , vol. 7 - pp e42688
Fogli A , Merle C, Roussel V, Schiffmann R, Ughetto S, Theisen M, Boespflug-Tanguy O
Developmental splicing deregulation in leukodystrophies related to EIF2B mutations.
Published on 01 Jan 2012 in PloS one , vol. 7 - pp e38264
Huyghe A, Horzinski L, Hénaut A, Gaillard M, Bertini E, Schiffmann R, Rodriguez D, Dantal Y, Boespflug-Tanguy O, Fogli A
Characterization of novel paternal ncRNAs at the Plagl1 locus, including Hymai, predicted to interact with regulators of active chromatin.
Published on 01 Jan 2012 in PloS one , vol. 7 - pp e38907
Iglesias-Platas I, Martin-Trujillo A, Cirillo D, Court F , Guillaumet-Adkins A, Camprubi C, Bourc'his D, Hata K, Feil R, Tartaglia G, Arnaud P , Monk D
Corneal transduction by intra-stromal injection of AAV vectors in vivo in the mouse and ex vivo in human explants.
Published on 01 Jan 2012 in PloS one , vol. 7 - pp e35318
Hippert C, Ibanes S, Serratrice N, Court F , Malecaze F, Kremer EJ, Kalatzis V
[Natural history of adult-onset eIF2B-related disorders: a multicentric survey of 24 cases].
Published on 30 Nov 2011 in Revue neurologique , vol. 167 - pp 802-11
Carra-Dalliere C, Horzinski L, Ayrignac X, Vukusic S, Rodriguez D, Mauguiere F, Peter L, Goizet C, Bouhour F, Denier C, Confavreux C, Obadia M, Blanc F, de Seze J, Sedel F, Guennoc AM, Sartori E, Laplaud D, Antoine JC, Fogli A , Boespflug-Tanguy O, Labauge P
Synergic reprogramming of mammalian cells by combined exposure to mitotic Xenopus egg extracts and transcription factors.
Published on 18 Oct 2011 in Proceedings of the National Academy of Sciences of the United States of America , vol. 108 - pp 17331-6
Ganier O, Bocquet S, Peiffer I, Brochard V, Arnaud P , Puy A, Jouneau A, Feil R, Renard JP, Méchali M
Long-range chromatin interactions at the mouse Igf2/H19 locus reveal a novel paternally expressed long non-coding RNA.
Published on 30 Aug 2011 in Nucleic acids research , vol. 39 - pp 5893-906
Court F , Baniol M, Hagege H, Petit JS, Lelay-Taha MN, Carbonell F, Weber M, Cathala G, Forne T
Human imprinted retrogenes exhibit non-canonical imprint chromatin signatures and reside in non-imprinted host genes.
Published on 30 Jun 2011 in Nucleic acids research , vol. 39 - pp 4577-86
Monk D, Arnaud P , Frost JM, Wood AJ, Cowley M, Martin-Trujillo A, Guillaumet-Adkins A, Iglesias Platas I, Camprubi C, Bourc'his D, Feil R, Moore GE, Oakey RJ
Molecular genetic analysis of the PLP1 gene in 38 families with PLP1-related disorders: identification and functional characterization of 11 novel PLP1 mutations.
Published on 16 Jun 2011 in Orphanet journal of rare diseases , vol. 6 - pp 40
Grossi S, Regis S, Biancheri R, Mort M, Lualdi S, Bertini E, Uziel G, Boespflug-Tanguy O, Simonati A, Corsolini F, Demir E, Marchiani V, Percesepe A, Stanzial F, Rossi A, Vaurs-Barrière C , Cooper DN, Filocamo M
Neurodegenerative disorder related to AIMP1/p43 mutation is not a PMLD.
Published on 11 Mar 2011 in American journal of human genetics , vol. 88 - pp 392-3; author reply 393-5
Boespflug-Tanguy O, Aubourg P, Dorboz I, Bégou M, Giraud G , Sarret C, Vaurs-Barrière C
Modulated contact frequencies at gene-rich loci support a statistical helix model for mammalian chromatin organization.
Published on 01 Jan 2011 in Genome biology , vol. 12 - pp R42
Court F , Miro J, Braem C, Lelay-Taha MN, Brisebarre A, Atger F, Gostan T, Weber M, Cathala G, Forné T
[Coupling proteinemia and serum protein electrophoresis: evaluation of the capillary technique (Capillarys 2, Sebia), experience from Clermont-Ferrand].
Published on 01 Nov 2010 in Annales de biologie clinique , vol. 68 - pp 657-67
Evaluation of the endoplasmic reticulum-stress response in eIF2B-mutated lymphocytes and lymphoblasts from CACH/VWM patients.
Published on 19 Oct 2010 in BMC neurology , vol. 10 - pp 94
Horzinski L, Kantor L, Huyghe A, Schiffmann R, Elroy-Stein O, Boespflug-Tanguy O, Fogli A
Genomic imprinting in germ cells: imprints are under control.
Published on 30 Sep 2010 in Reproduction (Cambridge, England) , vol. 140 - pp 411-23
Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment.
Published on 01 Aug 2010 in Development (Cambridge, England) , vol. 137 - pp 2483-92
Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P , Pombo A, O'Neill L, Azuara V
Genome-wide identification of new imprinted genes.
Published on 30 Jul 2010 in Briefings in functional genomics , vol. 9 - pp 304-14
Henckel A, Arnaud P
Peptidomics analysis of lymphoblastoid cell lines.
Published on 01 Jan 2010 in Methods in molecular biology (Clifton, N.J.) , vol. 615 - pp 247-57
Fogli A , Bulet P
Eukaryotic initiation factor 2B (eIF2B) GEF activity as a diagnostic tool for EIF2B-related disorders.
Published on 15 Dec 2009 in PloS one , vol. 4 - pp e8318
Horzinski L, Huyghe A, Cardoso MC, Gonthier C , Ouchchane L, Schiffmann R, Blanc P , Boespflug-Tanguy O, Fogli A
Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals.
Published on 15 Sep 2009 in Human molecular genetics , vol. 18 - pp 3375-83
Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P
Natural history of adult-onset eIF2B-related disorders: a multi-centric survey of 16 cases.
Published on 30 Aug 2009 in Brain : a journal of neurology , vol. 132 - pp 2161-9
Labauge P, Horzinski L, Ayrignac X, Blanc P , Vukusic S, Rodriguez D, Mauguiere F, Peter L, Goizet C, Bouhour F, Denier C, Confavreux C, Obadia M, Blanc F, de Sèze J, Fogli A , Boespflug-Tanguy O
Reciprocal imprinting of human GRB10 in placental trophoblast and brain: evolutionary conservation of reversed allelic expression.
Published on 15 Aug 2009 in Human molecular genetics , vol. 18 - pp 3066-74
Monk D, Arnaud P , Frost J, Hills FA, Stanier P, Feil R, Moore GE
Pelizaeus-Merzbacher-Like disease presentation of MCT8 mutated male subjects.
Published on 30 Jan 2009 in Annals of neurology , vol. 65 - pp 114-8
Vaurs-Barrière C , Deville M, Sarret C, Giraud G , Des Portes V, Prats-Viñas JM, De Michele G, Dan B, Brady AF, Boespflug-Tanguy O, Touraine R
A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10.
Published on 08 Oct 2008 in The EMBO journal , vol. 27 - pp 2523-32
Sanz LA, Chamberlain S, Sabourin JC, Henckel A, Magnuson T, Hugnot JP, Feil R, Arnaud P
Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing.
Published on 30 Aug 2008 in Genome research , vol. 18 - pp 1270-81
Monk D, Wagschal A, Arnaud P , Müller PS, Parker-Katiraee L, Bourc'his D, Scherer SW, Feil R, Stanier P, Moore GE
PLP1 splicing abnormalities identified in Pelizaeus-Merzbacher disease and SPG2 fibroblasts are associated with different types of mutations.
Published on 30 Aug 2008 in Human mutation , vol. 29 - pp 1028-36
Bonnet-Dupeyron MN, Combes P, Santander P, Cailloux F, Boespflug-Tanguy O, Vaurs-Barrière C
Absence of OLIG2 mutations in patients presenting with a severe Pelizaeus-Merzbacher-like leukodystrophy associated with motor neuron dysfunction.
Published on 05 Jun 2008 in American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics , vol. 147B - pp 538-9
Bonnet-Dupeyron MN, Combes P, Boespflug-Tanguy O, Vaurs-Barrière C
Sensitivity and specificity of decreased CSF asialotransferrin for eIF2B-related disorder.
Published on 03 Jun 2008 in Neurology , vol. 70 - pp 2226-32
Vanderver A, Hathout Y, Maletkovic J, Gordon ES, Mintz M, Timmons M, Hoffman EP, Horzinski L, Niel F, Fogli A , Boespflug-Tanguy O, Schiffmann R
Exon deletion in the non-catalytic domain of eIF2Bepsilon due to a splice site mutation leads to infantile forms of CACH/VWM with severe decrease of eIF2B GEF activity.
Published on 30 May 2008 in Annals of human genetics , vol. 72 - pp 410-5
Horzinski L, Gonthier C , Rodriguez D, Scherer C, Boespflug-Tanguy O, Fogli A
No evidence for association between the EIF2B5 gene and multiple sclerosis in French families.
Published on 30 May 2008 in Multiple sclerosis (Houndmills, Basingstoke, England) , vol. 14 - pp 573
Fogli A , Barbier C, Cournu-Rebeix I, Babron MC, Clerget-Darpoux F, Fontaine B, Boespflug-Tanguy O
Genes involved in leukodystrophies: a glance at glial functions.
Published on 30 May 2008 in Current neurology and neuroscience reports , vol. 8 - pp 217-29
Boespflug-Tanguy O, Labauge P, Fogli A , Vaurs-Barriere C